Quick and sensitive identification of multidrug-resistant germs
Researchers from the University of Basel have developed a sensitive testing system that allows the rapid and reliable detection of resistance in bacteria. The system is based on tiny, functionalized cantilevers that bend due to binding of sample material. In the analyses, the system was able to detect resistance in a sample quantity equivalent to 1–10 bacteria.
07 December 2020
Bacteria that are no longer susceptible to various antibiotics pose a significant threat to our health. In the event of a bacterial infection, physicians require rapid information about potential resistance so that they can respond quickly and correctly.
Cantilever systems as an alternative
Traditional methods for detecting resistance are based on cultivating bacteria and testing their sensitivity to a spectrum of antibiotics. These methods take 48 to 72 hours to deliver results, and some strains of bacteria are difficult to cultivate. Molecular biological tests are a great deal faster and work by amplifying resistance genes or specific short sequences of genetic material by polymerase chain reaction (PCR), but even this method doesn’t deliver satisfactory results for every bacterium.
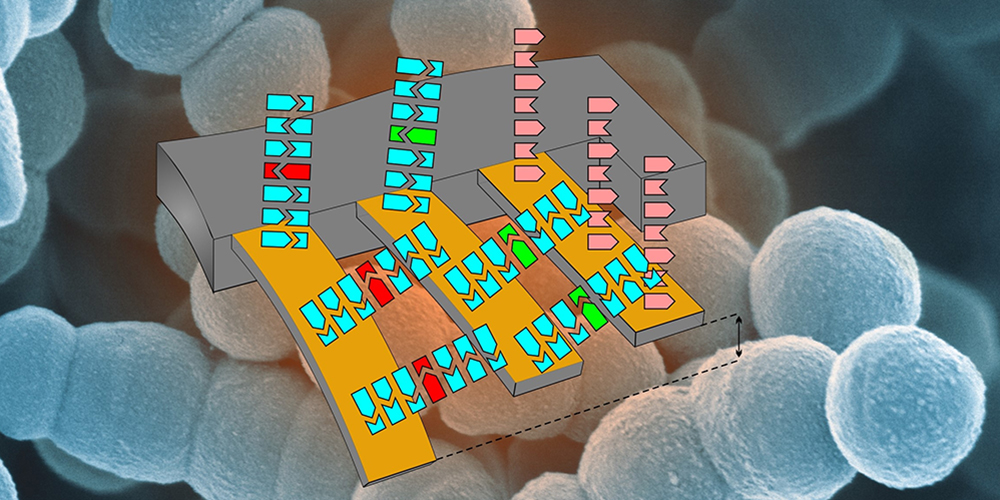
An alternative comes in the form of methods using tiny cantilevers, which bend when RNA molecules bind to their surface, for example — and this bending can then be detected. RNA molecules are “transcripts” of genes and can be used as instructions for building proteins. In addition, RNA molecules can be used to detect resistance genes in the genetic material of bacteria.
No need for labelling or amplification
Writing in the journal Global Challenges, a team of scientists from the Department of Physics, the Department of Biomedicine and the Swiss Nanoscience Institute (SNI) at the University of Basel have now presented a cantilever testing system that allowed them to detect RNA from a single antibiotic resistant bacterium. With the new cantilever system, it is not necessary to amplify or label the samples for analysis.
The researchers began by attaching sequences of three genes associated with vancomycin resistance to the cantilevers and then exposed these prepared cantilevers to a flow of RNA extracted from bacteria. If RNA molecules from the resistance genes were present, the matching RNA fragments would bind to the cantilevers, causing them to undergo nanoscale deflection that could be detected using a laser.
A clear signal even with point mutations
This method allowed the detection of not only resistance genes, but also individual point mutations associated with them. To study this, the researchers used point mutations coupled to genes responsible for resistance to ampicillin and other betalactam antibiotics.
“The big advantage of the method we’ve developed is its speed and sensitivity,” says Dr. François Huber, first author of the paper. “We succeeded in detecting tiny quantities of specific RNA fragments within five minutes.” In the case of single mutations, the detected RNA quantities corresponded to about 10 bacteria. When it came to detecting entire resistance genes, the researchers obtained a clear signal even with an amount of RNA that corresponded to a single bacterium.
“If we can detect specific genes or mutations in the genome of bacteria, then we know what antibiotic resistance the bacteria will exhibit,” explains Professor Adrian Egli from University Hospital Basel, whose team played an essential role in the study. “Our work in the hospital would benefit from this kind of reliable and sensitive information about the resistance of pathogens.”
Original publication
François Huber, Hans Peter Lang, Daniela Lang, Daniel Wüthrich, Vladimira Hinić, Christoph Gerber, Adrian Egli, Ernst Meyer
Rapid and ultrasensitive detection of mutations and genes relevant to antimicrobial resistance in bacteria
Global Challenges (2020), doi: 10.1002/gch2.202000066
Further information
Prof. Dr. Ernst Meyer, University of Basel Basel, Department of Physics, phone +41 61 207 37 24, email: ernst.meyer@unibas.ch
Dr. François Huber, University of Basel Basel, Department of Physics, phone +41 61 207 37 24, email: francois.huber@unibas.ch
Prof. Dr. Christoph Gerber, University of Basel Basel, Swiss Nanoscience Institute, Department of Physics, email: christoph.gerber@unibas.ch